Course materials for Quantitative Molecular Neuroscience Fall 2022 (ME.440.825.0001.FA22) from Stephanie Hicks.
Key resources
- Workshop material: pkgdown website
- Code: GitHub
Instructor
Stephanie C. Hicks, PhD
Associate Professor, Biostatistics, Johns Hopkins Bloomberg School of Public Health
Faculty member, Johns Hopkins Data Science Lab
Pronouns: she/her - web: https://www.stephaniehicks.com
- email: shicks19@jhu.edu
- twitter: @stephaniehicks
Acknowledgements
This course website was developed and is maintained by Stephanie C. Hicks.
The following individuals have contributed to improving the course or materials have been adapted from their courses:
- Mike Love
- Keegan Korthauer
- Orchestrating Single-Cell Analysis with Bioconductor (OSCA) contributors (cite: Amezquita et al. 2019)
The course materials are licensed under the Creative Commons Attribution 4.0 International License. Linked and embedded materials are governed by their own licenses. I assume that all external materials used or embedded here are covered under the educational fair use policy. If this is not the case and any material displayed here violates copyright, please let me know and I will remove it.
Create a copy of this repository
You can create a new GitHub repository in your own GitHub account that is a copy of this repository for yourself to edit/write code and notes. Github calls this “creating a template repository.” This is different than a fork because there will be no link between the original and your repository. From there, you can clone the repository with the git clone command and run / edit the code in the repository.
Creating a repository with the template can be done in the following way:
- Go to https://github.com/stephaniehicks/jhuquantneuro2022
- Click on the “Use this template” button at the top of the GitHub template repository
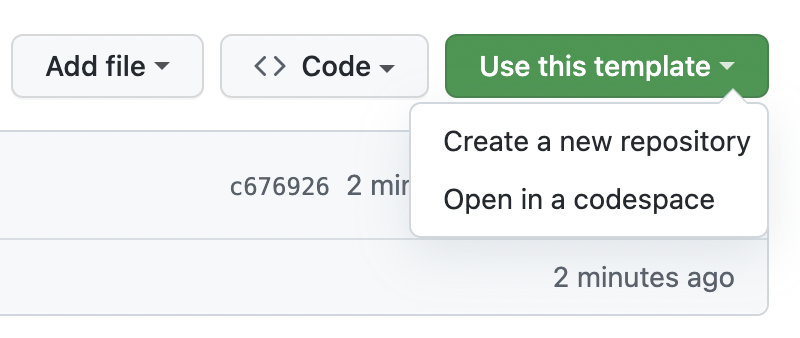
From there, you can follow these instructions. However do not optionally select “Include all branches” as you do not need this for your own projects.
Software
These materials use tidyverse and packages from Bioconductor version 3.15. This is the current ‘release’ version of Bioconductor, which can be installed following these instructions.
For example, you can then install a subset of the packages necessary for these tutorials using the following:
library(BiocManager)
install(c("SingleCellExperiment","scater", "scran"))Please see the DESCRIPTION for a full list of dependencies.