library(sloop)Introduction
Object oriented programming is one of the most successful and widespread philosophies of programming and is a cornerstone of many programming languages including Java, Ruby, Python, and C++.
At it’s core, object oriented programming (OOP) is a paradigm that is made up of classes and objects.
At a high-level, we use OOP to structure software programs into small, reusable pieces of code blueprints (i.e. classes), which are used to create instances of concrete objects.
The blueprint (or class) typically represents broad categories, e.g. bus or car that share attributes (e.g. color) (or fields).
- The classes specify what attributes you want, but not the actual values for a particular object.
- However, when you create instances with objects, you are specifying the attributes (e.g. a blue car, a red car, etc).
In addition, classes can also contain functions, called methods available only to objects of that type.
- These functions are defined within the class and perform some action helpful to that specific type of object.
- For example, our
carclass may have a methodrepaintthat changes the color attribute of our car. - This function is only helpful to objects of type
car, so we declare it within thecarclass thus making it a method.
OOP in R
Base R has three object oriented systems, because the roots of R date back to 1976, when the idea of object orientated programming was barely four years old.
New object oriented paradigms were added to R as they were invented, and they exist in their own R packages.
OOP is a little more challenging in R than in other languages because:
- There are multiple OOP systems to choose from. Here, I will focus on the following three: S3, R6, and S4.
- S3 and S4 are provided by base R (two older OOP languages).
- R6 is provided by the R6 package, and is similar to the
- Reference Classes, or RC for short, from base R. Programmers who are already familiar with object oriented programming will feel at home using RC.
There is disagreement about the relative importance of the OOP systems. Hadley Wickham thinks S3 is most important, followed by R6, then S4. Others believe that S4 is most important (e.g. Bioconductor community), followed by RC, and that S3 should be avoided. This means that different R communities use different systems.
S3 and S4 use generic function OOP which is rather different from the encapsulated OOP used by most languages popular today (the exception is Julia which also uses generic function OOP) (more on these later). Basically, while the underlying ideas of OOP are the same across languages, their expressions are rather different. This means that you can not immediately transfer your existing OOP skills to R.
Generally in R, functional programming is much more important than object-oriented programming, because you typically solve complex problems by decomposing them into simple functions, not simple objects.
This lesson focuses on the mechanics of OOP, not its effective use, and it may be challenging to fully understand if you have not done object-oriented programming before.
sloop
Before we go on I want to introduce the sloop package:
The sloop package (think “sail the seas of OOP”) provides a number of helpers that fill in missing pieces in base R. The first of these is sloop::otype(). It makes it easy to figure out the OOP system used by a wild-caught object:
otype(1:10)[1] "base"library(palmerpenguins)
otype(penguins)[1] "S3"mle_obj <- stats4::mle(function(x = 1) (x - 2) ^ 2)
otype(mle_obj)[1] "S4"OOP systems
Different people use OOP terms in different ways, so this section provides a quick overview of important vocabulary. The explanations are necessarily compressed, but we will come back to these ideas multiple times.
The main reason to use OOP is polymorphism (literally: many shapes).
- Polymorphism means that a developer can consider a function’s interface separately from its implementation, making it possible to use the same function form for different types of input.
- This is closely related to the idea of encapsulation: the user doesn’t need to worry about details of an object because they are encapsulated behind a standard interface.
To be concrete, polymorphism is what allows summary() to produce different outputs for numeric and factor variables:
summary(penguins$bill_length_mm) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
32.10 39.23 44.45 43.92 48.50 59.60 2 summary(penguins$species) Adelie Chinstrap Gentoo
152 68 124 You could imagine summary() containing a series of if-else statements, but that would mean only the original author could add new implementations. An OOP system makes it possible for any developer to extend the interface with implementations for new types of input.
To be more precise, OO systems call the type of an object its class, and an implementation for a specific class is called a method. Roughly speaking,
- a class defines what an object is and
- methods describe what that object can do
The class defines the fields (or attributes), the data possessed by every instance of that class. Classes are organised in a hierarchy so that if a method does not exist for one class, its parent’s method is used, and the child is said to inherit behavior.
- An ordered factor inherits from a regular factor.
- A generalized linear model inherits from a linear model.
The process of finding the correct method given a class is called method dispatch.
The two main paradigms of OOP differ in how methods and classes are related. We will call these paradigms encapsulated and functional:
In encapsulated OOP, methods belong to objects or classes, and method calls typically look like
object.method(arg1, arg2). This is called encapsulated because the object encapsulates both data (with fields) and behavior (with methods), and is the paradigm found in most popular languages.In functional OOP, methods belong to generic functions, and method calls look like ordinary function calls:
generic(object, arg2, arg3). This is called functional because from the outside it looks like a regular function call, and internally the components are also functions.
With this terminology in hand, we can now talk precisely about the different OO systems available in R.
OOP principles
Ok let’s talk more about some OOP principles. The first is is the idea of a class and an object.
The world is made up of physical objects - the chair you are sitting in, the clock next to your bed, the bus you ride every day, etc. Just like the world is full of physical objects, your programs can be made of objects as well.
A class is a blueprint for an object: it describes the parts of an object, how to make an object, and what the object is able to do.
If you were to think about a class for a bus (as in the public buses that roam the roads), this class would describe attributes for the bus like
- the number of seats on the bus
- the number of windows
- the color of the bus
- the top speed of the bus
- the maximum distance the bus can drive on one tank of gas
A method is a function that is associated with a class to perform an action.
Buses, in general, can perform the same actions, and these actions are also described in the class:
- a bus can open and close its doors
- the bus can steer
- the accelerator or the brake can be used to slow down or speed up the bus
A constructor is a method to specify attributes of the class to create a object with the specific attributes that we specified.
We will use the bus class in order to create individual bus objects.
To do this, we will create a constructor method for the bus class to return an individual bus object with the attributes that we specified.
If we want to make a new class that has all the same attributes and methods as an existing class, but also has additional attributes, we do not want to rewrite the entire class, but rather we want to define a new class that inherits from the original class.
Imagine that after making the bus class you might want to make a special kind of class for a party bus.
The party_bus class has all of the same attributes and methods as our bus class, but they also has additional attributes and methods like
- the number of refrigerators
- window blinds that can be opened and closed
- smoke machines that can be turned on and off
In this framework of inheritance, we talk about the bus class as the super-class of the party bus, and the party bus is the sub-class of the bus.
What this relationship means is that the party bus has all of the same attributes and methods as the bus class plus additional attributes and methods.
OOP in R (v2)
Base R provides three OOP systems: S3, S4, and reference classes (RC):
S3 is R’s first OOP system, and is described in Statistical Models in S. S3 is an informal implementation of functional OOP and relies on common conventions rather than ironclad guarantees. This makes it easy to get started with, providing a low cost way of solving many simple problems.
S4 is a formal and rigorous rewrite of S3, and was introduced in Programming with Data. It requires more upfront work than S3, but in return provides more guarantees and greater encapsulation. S4 is implemented in the base methods package, which is always installed with R.
You might wonder if S1 and S2 exist. They don’t: S3 and S4 were named according to the versions of S that they accompanied. The first two versions of S didn’t have any OOP framework.
- RC implements encapsulated OO. RC objects are a special type of S4 objects that are also mutable (i.e., instead of using R’s usual copy-on-modify semantics, they can be modified in place). This makes them harder to reason about, but allows them to solve problems that are difficult to solve in the functional OOP style of S3 and S4.
S3
S3 allows your functions to return rich results with user-friendly display and programmer-friendly internals.
S3 is used throughout base R, so it’s important to master if you want to extend base R functions to work with new types of input.
Conveniently everything in R is an object. By “everything” I mean every single “thing” in R including numbers, functions, strings, data frames, lists, etc.
And while everything in R is an object, not everything is object-oriented.
This confusion arises because the base objects come from S, and were developed before anyone thought that S might need an OOP system. The tools and nomenclature evolved organically over many years without a single guiding principle.
Most of the time, the distinction between objects and object-oriented objects is not important. But here we need to get into the nitty gritty details so we will use the terms base objects and OO objects to distinguish them.
[Source]
To tell the difference between a base and OO object, use is.object()
# A base object:
is.object(1:10)[1] FALSE# An OO object
is.object(mtcars)[1] TRUETechnically, the difference between base and OO objects is that OO objects have a “class” attribute:
attr(1:10, "class")NULLattributes(1:10)NULLattr(mtcars, "class")[1] "data.frame"attributes(mtcars)$names
[1] "mpg" "cyl" "disp" "hp" "drat" "wt" "qsec" "vs" "am" "gear"
[11] "carb"
$row.names
[1] "Mazda RX4" "Mazda RX4 Wag" "Datsun 710"
[4] "Hornet 4 Drive" "Hornet Sportabout" "Valiant"
[7] "Duster 360" "Merc 240D" "Merc 230"
[10] "Merc 280" "Merc 280C" "Merc 450SE"
[13] "Merc 450SL" "Merc 450SLC" "Cadillac Fleetwood"
[16] "Lincoln Continental" "Chrysler Imperial" "Fiat 128"
[19] "Honda Civic" "Toyota Corolla" "Toyota Corona"
[22] "Dodge Challenger" "AMC Javelin" "Camaro Z28"
[25] "Pontiac Firebird" "Fiat X1-9" "Porsche 914-2"
[28] "Lotus Europa" "Ford Pantera L" "Ferrari Dino"
[31] "Maserati Bora" "Volvo 142E"
$class
[1] "data.frame"This can be slightly confusing, but important to note:
You can find out the class of an object in R using the class() function, but the object may or may not have a class attribute.
class(1:10)[1] "integer"class("is in session.")[1] "character"class(mtcars)[1] "data.frame"class(class)[1] "function"Base types
While only OO objects have a class attribute, every object has a base type:
typeof(1:10)[1] "integer"typeof(mtcars)[1] "list"Base types do not form an OOP system because functions that behave differently for different base types are primarily written in C code that uses switch statements.
This means that only the R-core team can create new types, and creating a new type is a lot of work because every switch statement needs to be modified to handle a new case. As a consequence, new base types are rarely added.
In total, there are 25 different base types.
Here are some more base types we have already learned about:
typeof(NULL)[1] "NULL"typeof(1L)[1] "integer"typeof(1i)[1] "complex"OO objects
At a high-level, an S3 object is a base type with at least a class attribute.
Take the factor. Its base type is the integer vector, it has a class attribute of “factor”, and a levels attribute that stores the possible levels:
f <- factor(c("a", "b", "c"))
typeof(f)[1] "integer"attributes(f)$levels
[1] "a" "b" "c"
$class
[1] "factor"Let’s consider the penguins data frame.
- What is it’s base type?
- What is it’s
classattribute? - What other attributes does it have?
## try it hereCool. Let’s try creating a new class in the S3 system.
In the S3 system you can arbitrarily assign a class to any object. Class assignments can be made using the structure() function, or you can assign the class using class() and <-:
special_num_1 <- structure(1, class = "special_number")
special_num_1[1] 1
attr(,"class")
[1] "special_number"class(special_num_1)[1] "special_number"Let’s assign the number 2 to special_num_2 and look at the class of special_num_2.
### try it hereWhat’s happened here?
Next, let’s assign “special_number” to the class of special_num_2.
class(special_num_2) <- "special_number"
class(special_num_2)
special_num_2What’s happening here?
As crazy as this is, it is completely legal R code, but if you want to have a better behaved S3 class you should create a constructor which returns an S3 object.
Create a constructor called shape_s3()
Consider the shape_s3() function below, which is a constructor that returns a shape_S3 object:
shape_s3 <- function(side_lengths){
structure(list(side_lengths = side_lengths), class = "shape_S3")
}
square_4 <- shape_s3(c(4, 4, 4, 4))
class(square_4)[1] "shape_S3"triangle_3 <- shape_s3(c(3, 3, 3))
class(triangle_3)[1] "shape_S3"We have now made two shape_S3 objects: square_4 and triangle_3, which are both instantiations of the shape_S3 class.
Imagine that you wanted to create a method (or function) that would return TRUE if a shape_S3 object was a square, FALSE if a shape_S3 object was not a square, and NA if the object provided as an argument to the method was not a shape_S3 object.
This can be achieved using R’s generic methods system. A generic method can return different values based depending on the class of its input.
For example, mean() is a generic method that can find the average of a vector of number or it can find the “average day” from a vector of dates.
mean(c(2, 3, 7))[1] 4mean(c(as.Date("2016-09-01"), as.Date("2016-09-03")))[1] "2016-09-02"Create a generic method called is_square()
Now, let’s create a generic method for identifying shape_S3 objects that are squares.
UseMethod()
First, the creation of every generic method uses the UseMethod() function in the following way with only slight variations:
[name of method] <- function(x) UseMethod("[name of method]")Let’s call this method is_square:
is_square <- function(x) UseMethod("is_square")Next, we add the actual definition for the function to detect whether or not a shape is a square by specifying is_square.shape_S3.
By putting a dot (.) and then the name of the class after is_square, we can create a method that associates is_square with the shape_S3 class:
is_square.shape_S3 <- function(x){
length(x$side_lengths) == 4 &&
x$side_lengths[1] == x$side_lengths[2] &&
x$side_lengths[2] == x$side_lengths[3] &&
x$side_lengths[3] == x$side_lengths[4]
}
is_square(square_4)[1] TRUEis_square(triangle_3)[1] FALSESeems to be working well!
We also want is_square() to return NA when its argument is not a shape_S3.
We can specify is_square.default as a last resort if there is not method associated with the object passed to is_square().
is_square.default <- function(x){
NA
}
is_square("square")[1] NAis_square(c(1, 1, 1, 1))[1] NALet’s try printing square_4:
print(square_4)$side_lengths
[1] 4 4 4 4
attr(,"class")
[1] "shape_S3"Doesn’t that look ugly?
Create a generic method print() for shape_S3 class
Lucky for us print() is a generic method, so we can specify a print method for the shape_S3 class:
print.shape_S3 <- function(x){
if(length(x$side_lengths) == 3){
paste("A triangle with side lengths of", x$side_lengths[1],
x$side_lengths[2], "and", x$side_lengths[3])
} else if(length(x$side_lengths) == 4) {
if(is_square(x)){
paste("A square with four sides of length", x$side_lengths[1])
} else {
paste("A quadrilateral with side lengths of", x$side_lengths[1],
x$side_lengths[2], x$side_lengths[3], "and", x$side_lengths[4])
}
} else {
paste("A shape with", length(x$side_lengths), "sides.")
}
}
print(square_4)[1] "A square with four sides of length 4"print(triangle_3)[1] "A triangle with side lengths of 3 3 and 3"print(shape_s3(c(10, 10, 20, 20, 15)))[1] "A shape with 5 sides."print(shape_s3(c(2, 3, 4, 5)))[1] "A quadrilateral with side lengths of 2 3 4 and 5"Since printing an object to the console is one of the most common things to do in R, nearly every class has an associated print method!
To see all of the methods associated with a generic like print() use the methods() function:
length(methods(print))[1] 262head(methods(print), 10) [1] "print.acf" "print.activeConcordance"
[3] "print.AES" "print.anova"
[5] "print.aov" "print.aovlist"
[7] "print.ar" "print.Arima"
[9] "print.arima0" "print.AsIs" Inheritance
One last note on S3 with regard to inheritance.
In the previous section we discussed how a sub-class can inherit attributes and methods from a super-class.
Since you can assign any class to an object in S3, you can specify a super class for an object the same way you would specify a class for an object:
class(square_4)[1] "shape_S3"class(square_4) <- c("shape_S3", "square")
class(square_4)[1] "shape_S3" "square" To check if an object is a sub-class of a specified class you can use the inherits() function:
inherits(square_4, "square")[1] TRUEExample: S3 Class/Methods for Polygons
The S3 system doesn’t have a formal way to define a class but typically, we use a list to define the class and elements of the list serve as data elements.
Here is our definition of a polygon represented using Cartesian coordinates.
- The class contains an element called
xcoordandycoordfor the x- and y-coordinates, respectively. - The
make_poly()function is the “constructor” function for polygon objects. It takes as arguments a numeric vector of x-coordinates and a corresponding numeric vector of y-coordinates.
## Constructor function for polygon objects
## x a numeric vector of x coordinates
## y a numeric vector of y coordinates
make_poly <- function(x, y) {
if(length(x) != length(y))
stop("'x' and 'y' should be the same length")
## Create the "polygon" object
object <- list(xcoord = x, ycoord = y)
## Set the class name
class(object) <- "polygon"
object
}Now that we have a class definition, we can develop some methods for operating on objects from that class.
The first method that we will define is the print() method. The print() method should just show some simple information about the object and should not be too verbose—just enough information that the user knows what the object is.
Here the print() method just shows the user how many vertices the polygon has.
It is a convention for print() methods to return the object x invisibly using the invisible() function.
## Print method for polygon objects
## x an object of class "polygon"
print.polygon <- function(x, ...) {
cat("a polygon with", length(x$xcoord),
"vertices\n")
invisible(x)
}The invisible() function is useful when it is desired to have functions return values which can be assigned, but which do not print when they are not assigned.
These functions both return their argument
f1 <- function(x) x
f2 <- function(x) invisible(x)
f1(1) # prints[1] 1f2(1) # does not printHowever, when you assign the f2() function to an object, it does return the value
z <- f2(1)
z[1] 1Next is the summary() method.
The summary() method typically shows a bit more information and may even do some calculations, but does not print something. The general strategy of summary() methods is
- The
summary()method returns an object of class"summary_'class name'" - There is a separate
print()method for"summary_'class name'"objects.
For example, here is a summary() method for polygon objects that computes the ranges of the x- and y-coordinates.
## object an object of class "polygon"
summary.polygon <- function(object, ...) {
object <- list(rng.x = range(object$xcoord),
rng.y = range(object$ycoord))
class(object) <- "summary_polygon"
object
}The summary method simply returns an object of class summary_polygon.
Now the corresponding print() method for summary.polygon objects:
## Note: x an object of class "summary_polygon"
print.summary_polygon <- function(x, ...) {
cat("x:", x$rng.x[1], "-->", x$rng.x[2], "\n")
cat("y:", x$rng.y[1], "-->", x$rng.y[2], "\n")
invisible(x)
}Now we can make use of our new polygon class and methods (summary() and print()).
## Construct a new "polygon" object
x <- make_poly(1:4, c(1, 5, 2, 1))
attributes(x)$names
[1] "xcoord" "ycoord"
$class
[1] "polygon"We can use the print() to see what the object is.
print(x)a polygon with 4 verticesAnd we can use the summary() method to get a bit more information about the object.
out <- summary(x)
class(out)[1] "summary_polygon"print(out)x: 1 --> 4
y: 1 --> 5 Because of auto-printing we can just call the summary() method and let the results auto-print.
summary(x)x: 1 --> 4
y: 1 --> 5 From here, we could build other methods for interacting with our polygon object.
For example, it may make sense to define a plot() method or maybe methods for intersecting two polygons together.
S4
S4 is a rigorous system that forces you to think carefully about program design.
It’s particularly well-suited for building large systems that evolve over time and will receive contributions from many programmers. This is why it is used by the Bioconductor project, so another reason to learn S4 is to equip you to contribute to that project.
The S4 system is slightly more restrictive than S3, but it’s similar in many ways.
Constructors in S4
- To create a new class in S4 you need to use the
setClass()function. - You need to specify two (or three arguments) for this function:
Classwhich is the name of the class as a stringslots, which is a named list of attributes for the class with the class of those attributes specified- (optionally)
contains, which includes the super-class of they class you are specifying (if there is a super-class)
Take look at the class definition for a bus_S4 and a party_bus_S4 below:
setClass(Class = "bus_S4",
slots = list(n_seats = "numeric",
top_speed = "numeric",
current_speed = "numeric",
brand = "character"))
setClass(Class = "party_bus_S4",
slots = list(n_subwoofers = "numeric",
smoke_machine_on = "logical"),
contains = "bus_S4")Now that we have created the bus_S4 and the party_bus_S4 classes we can create bus objects using the new() function. The new() function’s arguments are the name of the class and values for each “slot” in our S4 object.
my_bus <- new("bus_S4", n_seats = 20, top_speed = 80,
current_speed = 0, brand = "Volvo")
my_busAn object of class "bus_S4"
Slot "n_seats":
[1] 20
Slot "top_speed":
[1] 80
Slot "current_speed":
[1] 0
Slot "brand":
[1] "Volvo"my_party_bus <- new("party_bus_S4", n_seats = 10, top_speed = 100,
current_speed = 0, brand = "Mercedes-Benz",
n_subwoofers = 2, smoke_machine_on = FALSE)
my_party_busAn object of class "party_bus_S4"
Slot "n_subwoofers":
[1] 2
Slot "smoke_machine_on":
[1] FALSE
Slot "n_seats":
[1] 10
Slot "top_speed":
[1] 100
Slot "current_speed":
[1] 0
Slot "brand":
[1] "Mercedes-Benz"You can use the @ operator to access the slots of an S4 object:
my_bus@n_seats[1] 20my_party_bus@top_speed[1] 100This is essentially the same as using the $ operator with a list or an environment.
Methods in S4
S4 classes use a generic method system that is similar to S3 classes. In order to implement a new generic method you need to use the setGeneric() function and the standardGeneric() function in the following way:
setGeneric("new_generic", function(x){
standardGeneric("new_generic")
})Let’s create a generic function called is_bus_moving() to see if a bus_S4 object is in motion:
setGeneric("is_bus_moving", function(x){
standardGeneric("is_bus_moving")
})[1] "is_bus_moving"Now, we need to actually define the function, which we can to with setMethod().
The setMethod() functions takes as arguments
- the name of the method as a string (or
f) - the method signature (
signature), which specifies the class of each argument for the method - the function definition of the method
setMethod(f = "is_bus_moving",
signature = c(x = "bus_S4"),
definition = function(x){
x@current_speed > 0
}
)
is_bus_moving(my_bus)[1] FALSEmy_bus@current_speed <- 1
is_bus_moving(my_bus)[1] TRUEIn addition to creating your own generic methods, you can also create a method for your new class from an existing generic.
First, use the setGeneric() function with the name of the existing method you want to use with your class, and then use the setMethod() function like in the previous example. Let’s make a print() method for the bus_S4 class:
setGeneric("print")[1] "print"setMethod(f = "print",
signature = c(x = "bus_S4"),
definition = function(x){
paste("This", x@brand, "bus is traveling at a speed of", x@current_speed)
})
print(my_bus)[1] "This Volvo bus is traveling at a speed of 1"print(my_party_bus)[1] "This Mercedes-Benz bus is traveling at a speed of 0"Reference Classes
With reference classes we leave the world of R’s old object oriented systems and enter the philosophies of other prominent object oriented programming languages. We can use the setRefClass() function to define a class’ fields, methods, and super-classes. Let’s make a reference class that represents a student:
Student <- setRefClass("Student",
fields = list(name = "character",
grad_year = "numeric",
credits = "numeric",
id = "character",
courses = "list"),
methods = list(
hello = function(){
paste("Hi! My name is", name)
},
add_credits = function(n){
credits <<- credits + n
},
get_email = function(){
paste0(id, "@jhu.edu")
}
))To recap: we have created a class definition called Student, which defines the student class. This class has five fields and three methods. To create a Student object use the new() method:
brooke <- Student$new(name = "Brooke", grad_year = 2019, credits = 40,
id = "ba123", courses = list("Ecology", "Calculus III"))
stephanie <- Student$new(name = "Stephanie", grad_year = 2021, credits = 10,
id = "shicks456", courses = list("Puppetry", "Elementary Algebra"))You can access the fields and methods of each object using the $ operator:
brooke$credits[1] 40stephanie$hello()[1] "Hi! My name is Stephanie"stephanie$get_email()[1] "shicks456@jhu.edu"Methods can change the state of an object, for instance in the case of the add_credits() function:
brooke$credits[1] 40brooke$add_credits(4)
brooke$credits[1] 44Notice that the add_credits() method uses the complex assignment operator (<<-). You need to use this operator if you want to modify one of the fields of an object with a method. You’ll learn more about this operator in the Expressions & Environments section.
Reference classes can inherit from other classes by specifying the contains argument when they’re defined. Let’s create a sub-class of Student called Grad_Student which includes a few extra features:
Grad_Student <- setRefClass("Grad_Student",
contains = "Student",
fields = list(thesis_topic = "character"),
methods = list(
defend = function(){
paste0(thesis_topic, ". QED.")
}
))
jeff <- Grad_Student$new(name = "Jeff", grad_year = 2021, credits = 8,
id = "jl55", courses = list("Fitbit Repair",
"Advanced Base Graphics"),
thesis_topic = "Batch Effects")
jeff$defend()[1] "Batch Effects. QED."Real-World Examples
Background
Continuous glucose monitoring (CGM) is novel sensor modality which estimates blood glucose quasi-continuously over 2 weeks in free-living conditions. This facilitates real-time management and comprehensive characterization of glucose for persons with diabetes.

OOP For CGM Data
CGM is often collected with other biomarkers, demographic information, and follow-up indicators when used in epidemiological studies. OOP makes it possible to store these different data modalities together in an Object for each person and to standardize operations upon a person’s data.
simulated_CGM <- as.numeric(arima.sim(model = list(ar = 0.8, ma = 0.5), n = 1440) + 80)
simulated_TS <- seq(from = Sys.time(), length.out = 1440, by = 15*60)
demo <- list(Age = 55, Gender = "F")
biomk <- list(BMI = 25, HbA1c = 8.5)S3 Constructor for CGM Object
cgm_s3 <- function(CGM_data, CGM_datetimes, demographics, biomarkers){
structure(list(Data = CGM_data, Times = CGM_datetimes,
Age = demographics$Age, Gender = demographics$Gender,
BMI = biomarkers$BMI, HbA1c = biomarkers$BMI), class = "cgm_S3")
}
cgm_profile <- cgm_s3(simulated_CGM, simulated_TS, demo, biomk)
class(cgm_profile)[1] "cgm_S3"S3 Methods for CGM Object
We may want to overwrite the generic “print” and “summary” methods, outputting a plot of the CGM data in the former case or returning a subset of pertinent information in the latter.
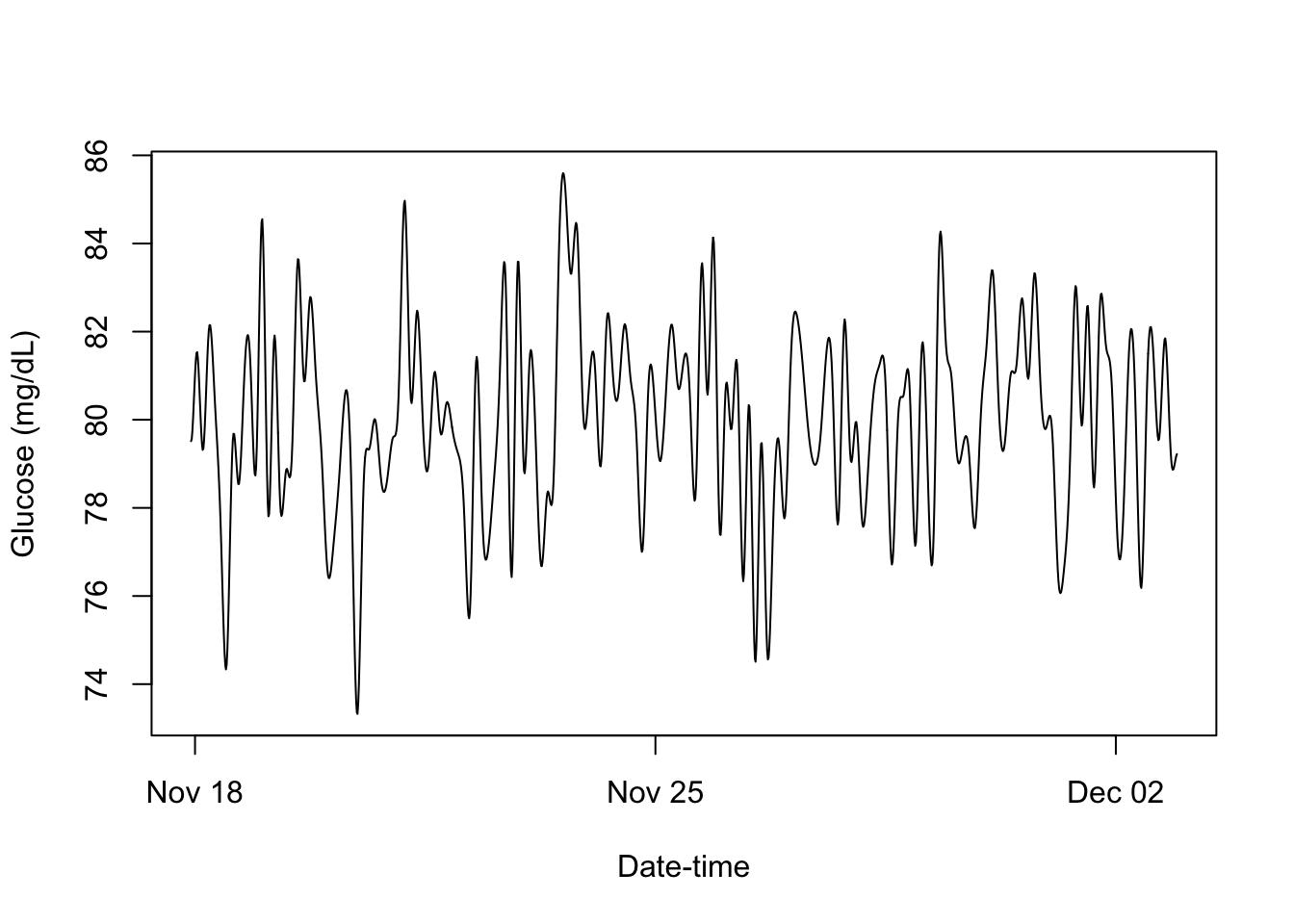
print.cgm_S3 <- function(x, ...){
plot(x = x$Times, y = x$Data, type = "l",
xlab = "Date-time", ylab = "Glucose (mg/dL)")
invisible(x)
}
print(cgm_profile)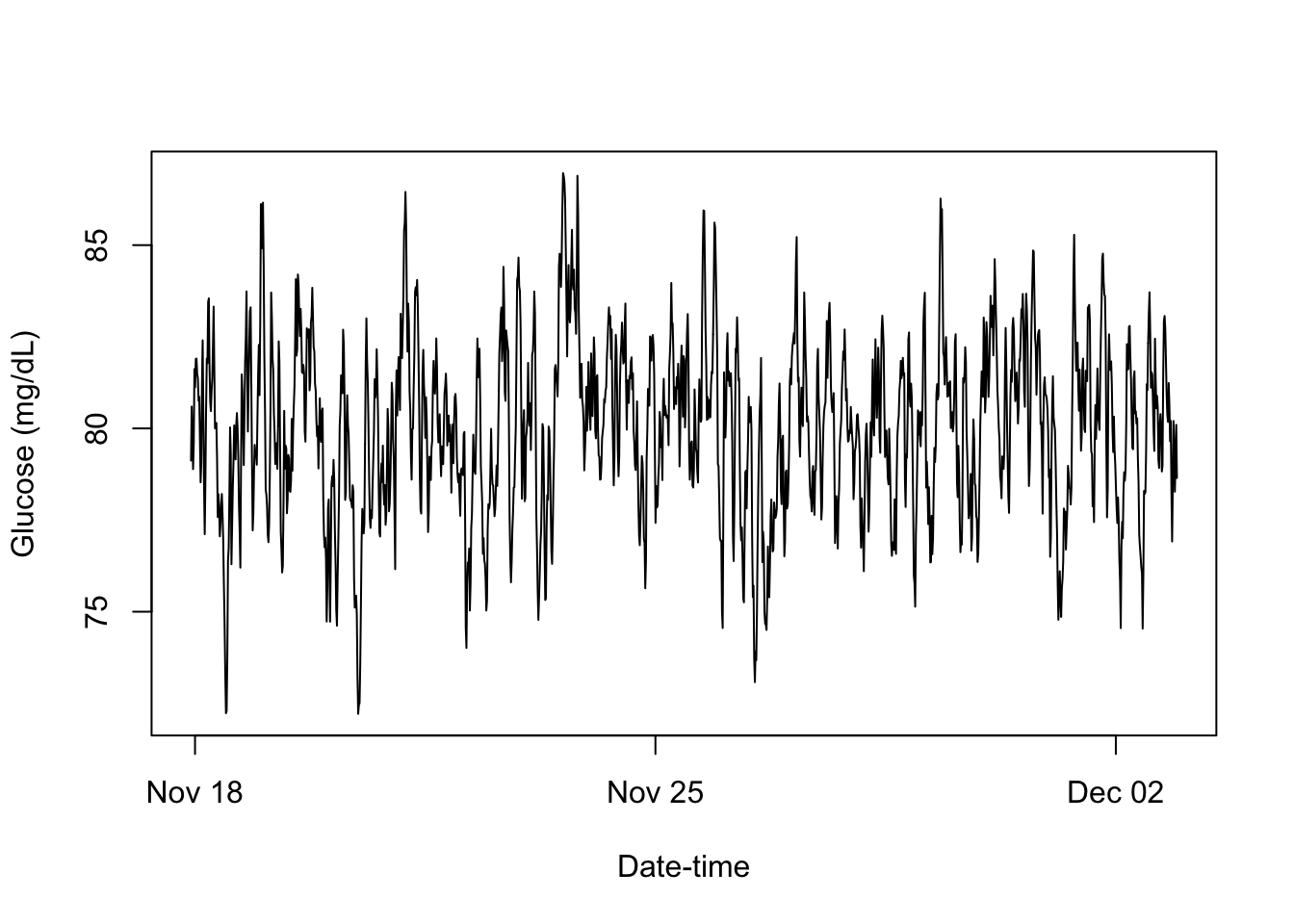
summary.cgm_S3 <- function(x, ...){
demo_string <- paste0("Age: ", x$Age, ", Gender: ", x$Gender)
biomk_string <- paste0("BMI: ", x$BMI, ", HbA1c: ", x$HbA1c)
cgm_string <- paste0("Mean Glucose: ", round(mean(x$Data), 2), ", Std. Dev. Glucose: ", round(sd(x$Data), 2))
object <- list(demographics = demo_string,
biomarkers = biomk_string,
cgm = cgm_string)
class(object) <- "summary_CGM"
object
}
print.summary_CGM <- function(x, ...){
cat(paste(paste0("Demographics - ", x$demographics),
paste0("Biomarkers - ", x$biomarkers),
paste0("CGM Metrics - ", x$cgm), sep = "\n"))
invisible(x)
}
summary(cgm_profile)Demographics - Age: 55, Gender: F
Biomarkers - BMI: 25, HbA1c: 25
CGM Metrics - Mean Glucose: 80, Std. Dev. Glucose: 2.37We may also wish to perform any range of standard operations upon the CGM time series data, like smoothing.
smooth <- function(x) UseMethod("smooth")
smooth.cgm_S3 <- function(x){
smoothed_CGM <- fitted(lm(x$Data ~ bs(x$Times, df = 12*14)))
x$Data <- smoothed_CGM
return(x)
}
smoothed_cgm_profile = smooth(cgm_profile)
print(smoothed_cgm_profile)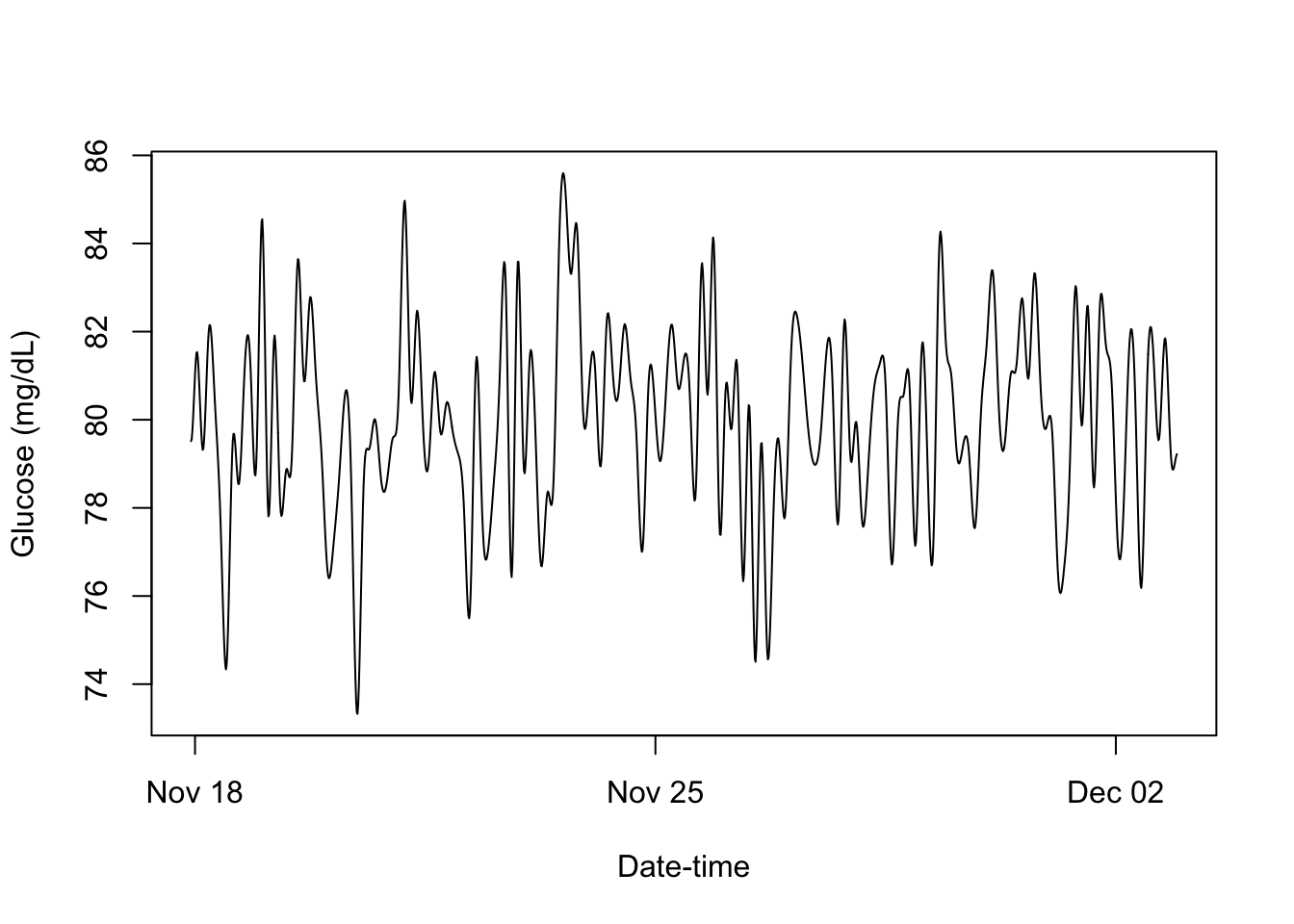
S4 Constructor for CGM Object
setClass(Class = "cgm_S4",
slots = list(Data = "numeric",
Times = "POSIXct",
Age = "numeric",
Gender = "character",
BMI = "numeric",
HbA1c = "numeric"))
cgm_profile <- new("cgm_S4", Data = simulated_CGM, Times = simulated_TS,
Age = demo$Age, Gender = demo$Gender, BMI = biomk$BMI,
HbA1c = biomk$HbA1c)
class(cgm_profile)[1] "cgm_S4"
attr(,"package")
[1] ".GlobalEnv"S4 Methods for CGM Object
We can overwrite the generic “print” and “summary” methods to achieve the same functionality in S4.
setGeneric("print")[1] "print"setGeneric("summary")[1] "summary"setGeneric("smooth", function(x){
standardGeneric("smooth")
})[1] "smooth"setMethod(f = "print",
signature = c(x = "cgm_S4"),
definition = function(x){
plot(x = x@Times, y = x@Data, type = "l",
xlab = "Date-time", ylab = "Glucose (mg/dL)")
invisible(x)
})
print(cgm_profile)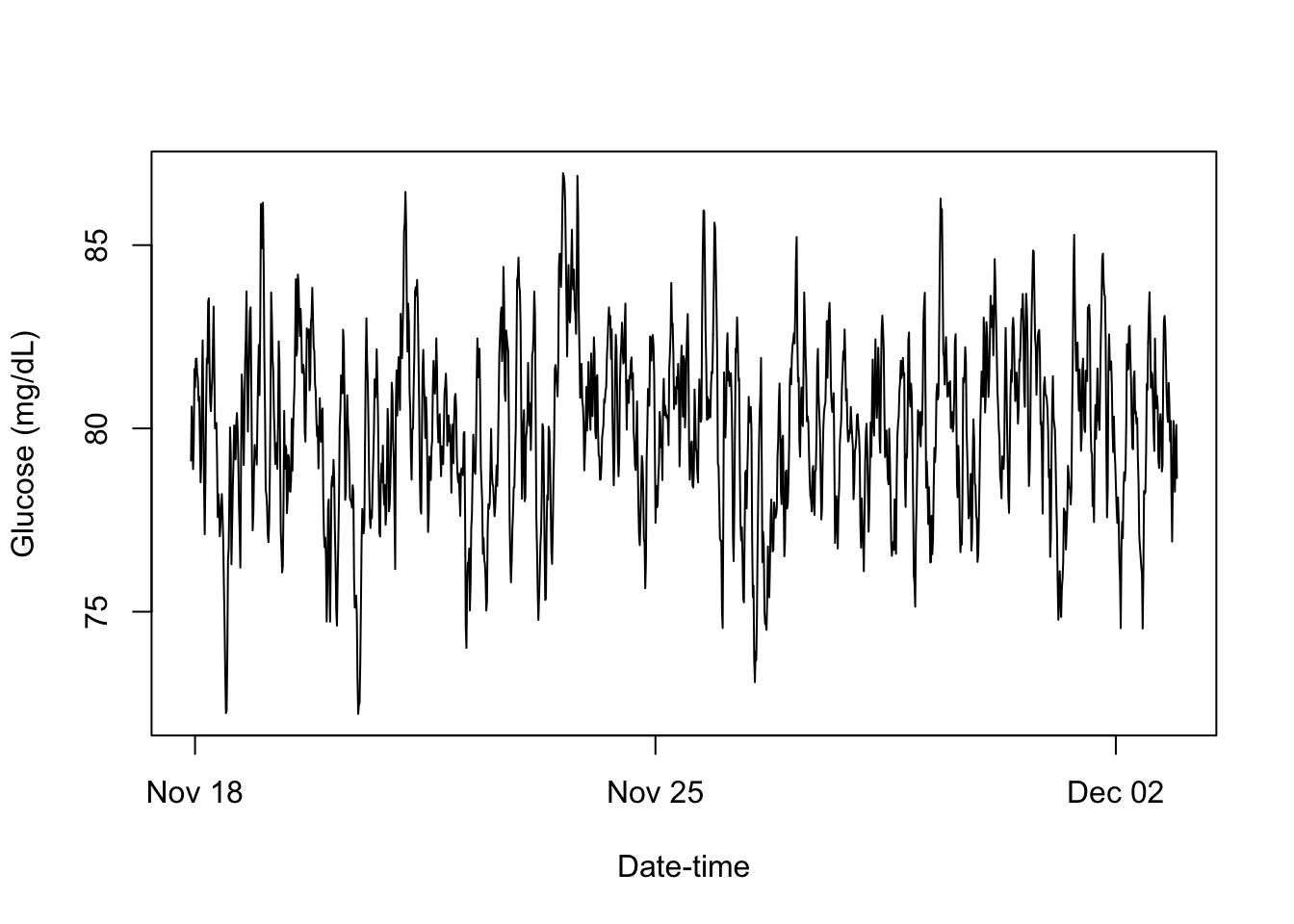
setMethod(f = "summary",
signature = c(object = "cgm_S4"),
definition = function(object){
MG = round(mean(object@Data), 2)
SDG = round(sd(object@Data), 2)
demo_string <- paste0("Demographics - Age: ", object@Age, ", Gender: ", object@Gender)
biomk_string <- paste0("Biomarkers - BMI: ", object@BMI, ", HbA1c: ", object@HbA1c)
cgm_string <- paste0("CGM Metrics - Mean Glucose: ", MG, ", Std. Dev. Glucose: ", SDG)
cat(paste(demo_string, biomk_string, cgm_string, sep = "\n"))
invisible(object)
})
summary(cgm_profile)Demographics - Age: 55, Gender: F
Biomarkers - BMI: 25, HbA1c: 8.5
CGM Metrics - Mean Glucose: 80, Std. Dev. Glucose: 2.37setMethod(f = "smooth",
signature = c(x = "cgm_S4"),
definition = function(x){
smoothed_CGM <- fitted(lm(x@Data ~ bs(x@Times, df = 12*14)))
x@Data <- smoothed_CGM
x
})
smoothed_cgm_profile = smooth(cgm_profile)
print(smoothed_cgm_profile)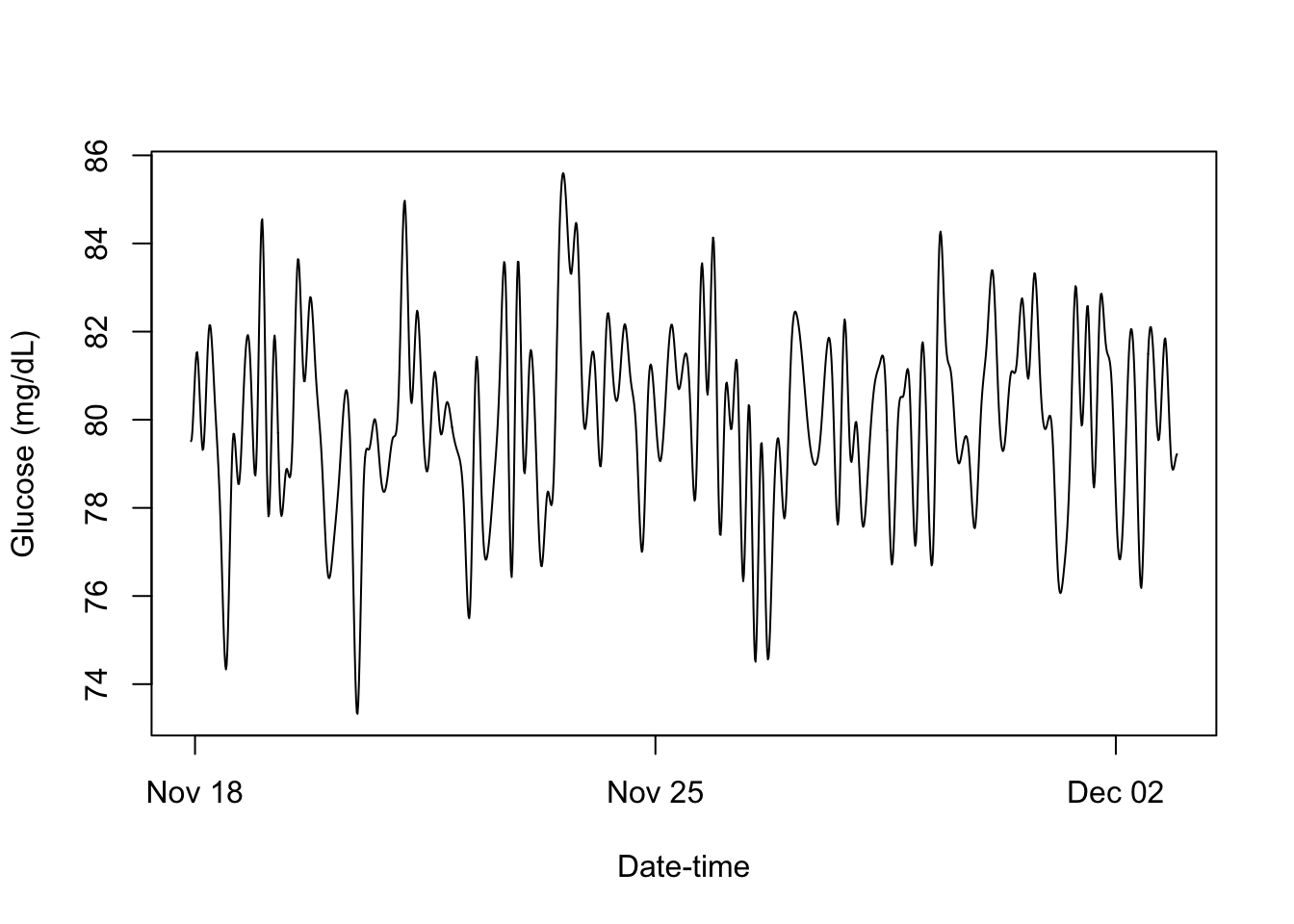
RC Constructor for CGM Object with Methods
All of the same functionality can also be replicated using Reference Classes. This alternative framework might even be preferable for this application, as packaging methods with objects compartmentalizes functionality for easily-distributable and accessible software. The objects are also mutable, unlike S3 and S4, which is preferable for certain types of objects which should be fluid when used.
cgm_RC <- setRefClass("cgm_RC",
fields = list(Data = "numeric",
Times = "POSIXct",
Age = "numeric",
Gender = "character",
BMI = "numeric",
HbA1c = "numeric"),
methods = list(
print = function(){
plot(x = Times, y = Data, type = "l",
xlab = "Date-time", ylab = "Glucose (mg/dL)")
invisible(.self)
},
summmary = function(){
MG = round(mean(Data), 2)
SDG = round(sd(Data), 2)
demo_string <- paste0("Demographics - Age: ", Age, ", Gender: ", Gender)
biomk_string <- paste0("Biomarkers - BMI: ", BMI, ", HbA1c: ", HbA1c)
cgm_string <- paste0("CGM Metrics - Mean Glucose: ", MG, ", Std. Dev. Glucose: ", SDG)
cat(paste(demo_string, biomk_string, cgm_string, sep = "\n"))
invisible(.self)
},
smooth = function(){
smoothed_CGM <- fitted(lm(Data ~ bs(Times, df = 12*14)))
Data <<- smoothed_CGM
}
))
cgm_profile <- cgm_RC$new(Data = simulated_CGM, Times = simulated_TS,
Age = demo$Age, Gender = demo$Gender, BMI = biomk$BMI,
HbA1c = biomk$HbA1c)cgm_profile$print()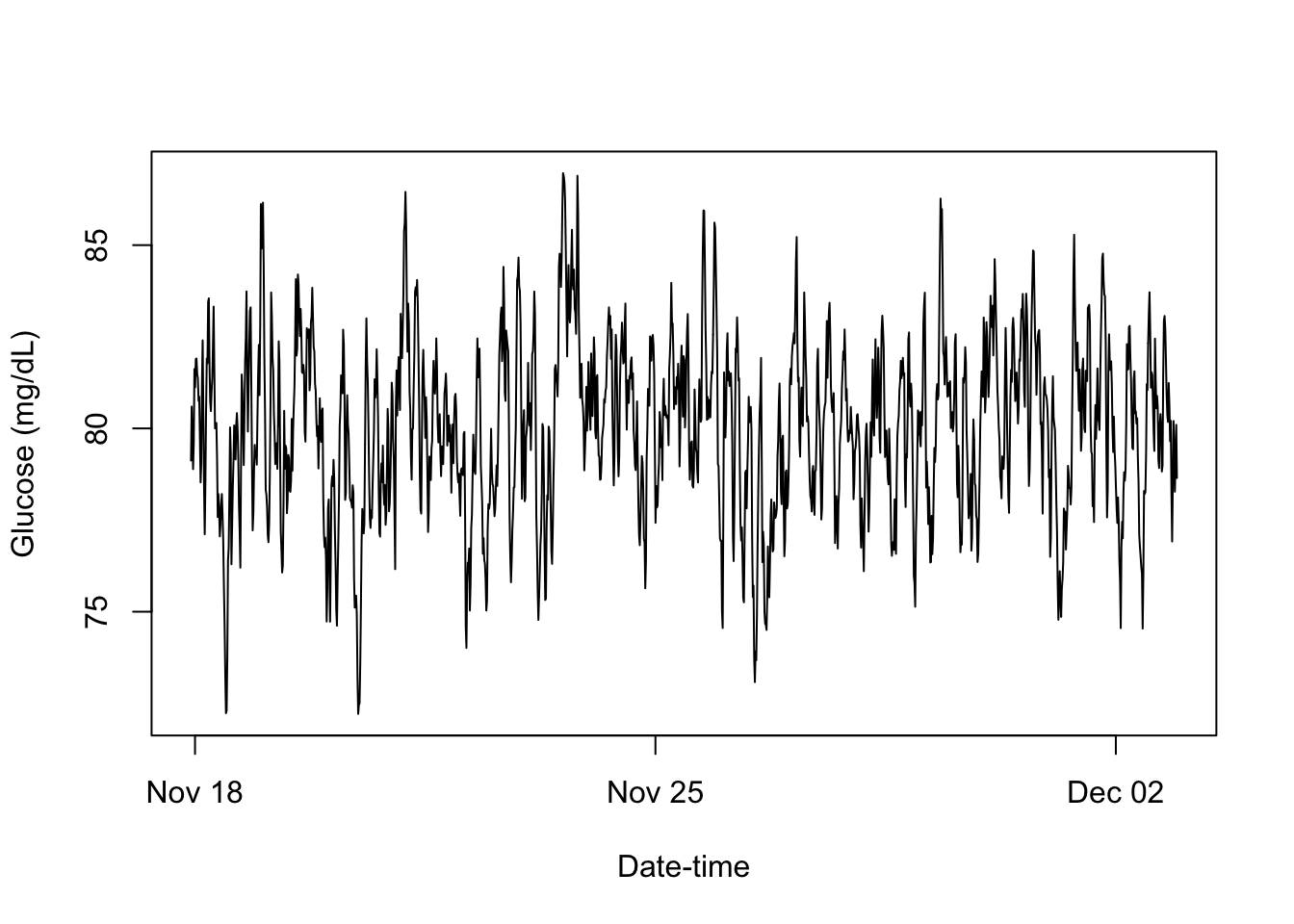
cgm_profile$summmary()Demographics - Age: 55, Gender: F
Biomarkers - BMI: 25, HbA1c: 8.5
CGM Metrics - Mean Glucose: 80, Std. Dev. Glucose: 2.37cgm_profile$smooth()
cgm_profile$print()