library(tidyverse)
library(DBI)
library(RSQLite)
library(dbplyr)Installation
We will load them here before kicking off the lecture.
Relational databases
Data live anywhere and everywhere. Data might be stored simply in a .csv or .txt file.
Data might be stored in an Excel or Google Spreadsheet. Data might be stored in large databases that require users to write special functions to interact with to extract the data they are interested in.
A relational database is a digital database based on the relational model of data, as proposed by E. F. Codd in 1970.
Broadly relational databases are a way to store and manipulate information.
- Databases are arranged as tables with a unique relation variable name
- Each table has columns (also known as fields or attributes) that describe the data
- Rows (also known as records) contain the data

[Source: Wikipedia]
When we are using a spreadsheet, we put formulas into cells to calculate new values based on old ones.
When we are using a database, we send commands (usually called queries) to a database manager (a program that manipulates the database for us).
The database manager does whatever lookups and calculations the query specifies, returning the results in a tabular form that we can then use as a starting point for further queries.
Examples of database manager include:
- Oracle
- IBM DB2
- PostgreSQL
- MySQL
- Microsoft Access
- SQLite (which is what we will use here)
A system used to maintain relational databases is a relational database management system (RDBMS).
Languages
We write queries in a language called Structured Query Language (SQL), which provides hundreds of different ways to analyze and recombine data.
Many database managers understand SQL but each stores data in a different way, so a database created with one cannot be used directly by another.
However, every database manager can import and export data in a variety of formats like .csv, .sql, so it is possible to move information from one to another.
Next, we will some example SQL queries that are common tasks for data scientists.
SQL basics
In this next few sections, we will use the SQLite database manager and interact with it interactively on the command-line with the command sqlite3.
In order to use the SQLite commands interactively, we need to enter into the SQLite console. So, open up a terminal, and run
Bash
cd data
sqlite3 survey.dbThe SQLite command is sqlite3 and you are telling SQLite to open up the survey.db. You need to specify the .db file, otherwise SQLite will open up a temporary, empty database.
- All SQLite-specific commands are prefixed with a
.to distinguish them from SQL commands. - To get out of SQLite, type out
.exitor.quit. For some terminals,Ctrl-Dcan also work.
- If you forget any SQLite
.(dot) command, type.help.
Example data
Before we get into using SQLite to select the data, let’s take a look at the tables of the database we will use in our examples:
Person: People who took readings, id being the unique identifier for that person.
| id | personal | family |
|---|---|---|
| dyer | William | Dyer |
| pb | Frank | Pabodie |
| lake | Anderson | Lake |
| roe | Valentina | Roerich |
| danforth | Frank | Danforth |
Site: Locations of the sites where readings were taken.
| name | lat | long |
|---|---|---|
| DR-1 | -49.85 | -128.57 |
| DR-3 | -47.15 | -126.72 |
| MSK-4 | -48.87 | -123.4 |
Visited: Specific identification id of the precise locations where readings were taken at the sites and dates.
| id | site | dated |
|---|---|---|
| 619 | DR-1 | 1927-02-08 |
| 622 | DR-1 | 1927-02-10 |
| 734 | DR-3 | 1930-01-07 |
| 735 | DR-3 | 1930-01-12 |
| 751 | DR-3 | 1930-02-26 |
| 752 | DR-3 | -null- |
| 837 | MSK-4 | 1932-01-14 |
| 844 | DR-1 | 1932-03-22 |
Survey: The measurements taken at each precise location on these sites. They are identified as taken. The field quant is short for quantity and indicates what is being measured. The values are rad, sal, and temp referring to ‘radiation’, ‘salinity’ and ‘temperature’, respectively.
| taken | person | quant | reading |
|---|---|---|---|
| 619 | dyer | rad | 9.82 |
| 619 | dyer | sal | 0.13 |
| 622 | dyer | rad | 7.8 |
| 622 | dyer | sal | 0.09 |
| 734 | pb | rad | 8.41 |
| 734 | lake | sal | 0.05 |
| 734 | pb | temp | -21.5 |
| 735 | pb | rad | 7.22 |
| 735 | -null- | sal | 0.06 |
| 735 | -null- | temp | -26.0 |
| 751 | pb | rad | 4.35 |
| 751 | pb | temp | -18.5 |
| 751 | lake | sal | 0.1 |
| 752 | lake | rad | 2.19 |
| 752 | lake | sal | 0.09 |
| 752 | lake | temp | -16.0 |
| 752 | roe | sal | 41.6 |
| 837 | lake | rad | 1.46 |
| 837 | lake | sal | 0.21 |
| 837 | roe | sal | 22.5 |
| 844 | roe | rad | 11.25 |
SQL .tables and .schema
In an interactive sqlite3 session,
- Type
.tablesto list the tables in the database - Type
.schemato see the SQL statements used to create the tables in the database. The statements will have a list of the columns and the data types each column stores.
.schema
The output from .schema is formatted as <columnName dataType>.
Output
CREATE TABLE Person (id text, personal text, family text);
CREATE TABLE Site (name text, lat real, long real);
CREATE TABLE Survey (taken integer, person text, quant text, reading real);
CREATE TABLE Visited (id integer, site text, dated text);Thus we can see from the first line that the table Person has three columns:
- id with type text
- personal with type text
- family with type text
The available data types vary based on the database manager - you can search online for what data types are supported.
SQL command SELECT
For now, let’s write an SQL query that displays scientists’ names.
We do this using the SQL command SELECT, giving it the names of the columns we want and the table we want them from.
Our query looks like this:
SQL
SELECT family, personal FROM Person;And the output looks like this:
Output
|family |personal |
|--------|---------|
|Dyer |William |
|Pabodie |Frank |
|Lake |Anderson |
|Roerich |Valentina|
|Danforth|Frank |The semicolon at the end of the query tells the database manager that the query is complete and ready to run.
We have written our commands in upper case and the names for the table and columns in lower case, but we don’t have to because SQL is case insensitive.
SQL
SeLeCt FaMiLy, PeRsOnAl FrOm PeRsOn;Output:
Output
|family |personal |
|--------|---------|
|Dyer |William |
|Pabodie |Frank |
|Lake |Anderson |
|Roerich |Valentina|
|Danforth|Frank |You can use SQL’s case insensitivity to distinguish between different parts of an SQL statement.
Here, we use the convention of using
- UPPER CASE for SQL keywords (such as
SELECTandFROM) - Title Case for table names
- lower case for field names
Whatever casing convention you choose, please be consistent: complex queries are hard enough to read without the extra cognitive load of random capitalization.
;
While we are on the topic of SQL’s syntax, one aspect of SQL’s syntax that can frustrate novices and experts alike is forgetting to finish a command with ; (semicolon).
When you press enter for a command without adding the ; to the end, it can look something like this:
Output
SELECT id FROM Person
...>
...>This is SQL’s prompt, where it is waiting for additional commands or for a ; to let SQL know to finish.
This is easy to fix! Just type ; and press enter!
More about SELECT
Row and columns in a database table are not actually stored in any particular order.
They will always be displayed in some order, but we can control that in various ways.
We could swap the columns in the output by writing our query as:
SQL
SELECT personal, family FROM Person;Output
|personal |family |
|---------|--------|
|William |Dyer |
|Frank |Pabodie |
|Anderson |Lake |
|Valentina|Roerich |
|Frank |Danforth|or even repeat columns:
SQL
SELECT id, id, id FROM Person;Output
|id |id |id |
|--------|--------|--------|
|dyer |dyer |dyer |
|pb |pb |pb |
|lake |lake |lake |
|roe |roe |roe |
|danforth|danforth|danforth|The * operator
As a shortcut, we can select all of the columns in a table using *:
SQL
SELECT * FROM Person;Output
|id |personal |family |
|--------|---------|--------|
|dyer |William |Dyer |
|pb |Frank |Pabodie |
|lake |Anderson |Lake |
|roe |Valentina|Roerich |
|danforth|Frank |Danforth|Sorting and removing duplicates
In this section, we will explore the following questions of the Antarctic data
- What are the unique types of measurements taken in
Survey? - Which scientists took measurements on the expedition?
To answer the first question, we will extract the values in column quant (short for quantity) from Survey, which contains values rad, sal, and temp referring to ‘radiation’, ‘salinity’ and ‘temperature’, respectively.
However, we only want the unique value labels.
The following will extract the quant column from the Survey table, but not return unique / distinct labels.
SQL
SELECT quant FROM Survey;But, adding the DISTINCT keyword to our query eliminates the redundant output to make the result more readable:
SQL
SELECT DISTINCT quant FROM Survey;Output
|quant|
|-----|
|rad |
|sal |
|temp |You can also use the DISTINCT keyword on multiple columns.
If we select more than one column, distinct sets of values are returned (in this case pairs, because we are selecting two columns) and duplicates are removed:
SQL
SELECT DISTINCT taken, quant FROM Survey;Output
|taken|quant|
|-----|-----|
|619 |rad |
|619 |sal |
|622 |rad |
|622 |sal |
|734 |rad |
|734 |sal |
|734 |temp |
|735 |rad |
|735 |sal |
|735 |temp |
|751 |rad |
|751 |temp |
|751 |sal |
|752 |rad |
|752 |sal |
|752 |temp |
|837 |rad |
|837 |sal |
|844 |rad |Next, we will look at the Person table and sort the scientists names.
Database records are not necessarily sorted in any particular order.
If you want to have the table returned sorted in a particular way, you add the ORDER BY clause to our query:
SQL
SELECT * FROM Person ORDER BY id;Output
|id |personal |family |
|-------|---------|--------|
|danfort|Frank |Danforth|
|dyer |William |Dyer |
|lake |Anderson |Lake |
|pb |Frank |Pabodie |
|roe |Valentina|Roerich |The default is to sort in an ascending order, but we can sort in a descending order using DESC (for “descending”):
SQL
SELECT * FROM person ORDER BY id DESC;Output
|id |personal |family |
|-------|---------|--------|
|roe |Valentina|Roerich |
|pb |Frank |Pabodie |
|lake |Anderson |Lake |
|dyer |William |Dyer |
|danfort|Frank |Danforth|(And if we want to make it clear that we’re sorting in ascending order, we can use ASC instead of DESC.)
Let’s look at which scientist (person) measured what quantities (quant) during each visit (taken) with the Survey table.
We also want to sort by two columns at once
- Sort results first in ascending order by
taken - And then in descending order by
personwithin each group of equaltakenvalues:
SQL
SELECT taken, person, quant FROM Survey ORDER BY taken ASC, person DESC;Output
|taken|person|quant|
|-----|------|-----|
|619 |dyer |rad |
|619 |dyer |sal |
|622 |dyer |rad |
|622 |dyer |sal |
|734 |pb |rad |
|734 |pb |temp |
|734 |lake |sal |
|735 |pb |rad |
|735 |-null-|sal |
|735 |-null-|temp |
|751 |pb |rad |
|751 |pb |temp |
|751 |lake |sal |
|752 |roe |sal |
|752 |lake |rad |
|752 |lake |sal |
|752 |lake |temp |
|837 |roe |sal |
|837 |lake |rad |
|837 |lake |sal |
|844 |roe |rad |This query gives us a good idea of which scientist was involved in which visit, and what measurements they performed during the visit.
Looking at the table, it seems like some scientists specialized in certain kinds of measurements.
We can examine which scientists performed which measurements by selecting the appropriate columns and removing duplicates.
SQL
SELECT DISTINCT quant, person FROM Survey ORDER BY quant ASC;Output
|quant|person|
|-----|------|
|rad |dyer |
|rad |pb |
|rad |lake |
|rad |roe |
|sal |dyer |
|sal |lake |
|sal |-null-|
|sal |roe |
|temp |pb |
|temp |-null-|
|temp |lake |Other important tasks
There are many other tasks you can do with SQL, but for purposes of the lecture, I will leave you to work through this carpentries tutorial if you want to know more:
Filtering
How can you select subsets of data? You use WHERE.
Here is an example of filtering for all rows that contain “dyer” in the Person column.
SQL
SELECT * FROM Survey WHERE person = "dyer";Output
619|dyer|rad|9.82
619|dyer|sal|0.13
622|dyer|rad|7.8
622|dyer|sal|0.09For more information about filtering, read through this tutorial:
And more
The carpentries tutorial has so much more including how to:
- Calculating new values (https://swcarpentry.github.io/sql-novice-survey/04-calc)
- How to deal with missing data (https://swcarpentry.github.io/sql-novice-survey/05-null)
- How to aggregate data to calculate summaries (https://swcarpentry.github.io/sql-novice-survey/06-agg)
- How to write queries that joins together two tables (https://swcarpentry.github.io/sql-novice-survey/07-join)
- How to create tables or modify exisiting data in tables (https://swcarpentry.github.io/sql-novice-survey/09-create)
SQL in R
Thus far, everything we have done with SQL has been through an interactive session with sqlite3.
You can also access a database with R (and other programming languages too!). Library and functions may differ, but concepts are the same.
Connect to the SQL database
The main workhorse packages that we will use are the DBI and RSQLite packages.
DBIis an R package that connects R to database management systems (DBMS).DBIseparates the connectivity to the DBMS into a “front-end” and a “back-end”. The package defines an interface that is implemented by DBI backends such as RPostgres, RMariaDB, RSQLite, odbc, bigrquery, and more!RSQLiteis an R package that embeds the SQLite database engine in R, providing a DBI-compliant interface.SQLiteis a public-domain, single-user, very light-weight database engine that implements a decent subset of the SQL 92 standard, including the core table creation, updating, insertion, and selection operations, plus transaction management.
Example workflow
Here’s a short R program that sorts the scientists names in a descending order from from an SQLite database stored in a file called survey.db:
library(RSQLite)
connection <- dbConnect(drv = RSQLite::SQLite(),
dbname = here::here("readings","05-database-programming", "data", "survey.db"))
results <- dbGetQuery(connection, "SELECT * FROM Person ORDER BY id DESC;")
print(results) id personal family
1 roe Valentina Roerich
2 pb Frank Pabodie
3 lake Anderson Lake
4 dyer William Dyer
5 danforth Frank DanforthdbDisconnect(connection)Let’s break this down.
The program starts by importing the RSQLite library.
If we were connecting to MySQL, DB2, or some other database, we would import a different library, but all of them provide the same functions, so that the rest of our program does not have to change (at least, not much) if we switch from one database to another.
Line 2 establishes a connection to the database.
Since we’re using SQLite, all we need to specify is the name of the database file. Other systems may require us to provide a username and password as well.
On line 3, we retrieve the results from an SQL query.
It’s our job to make sure that SQL is properly formatted; if it isn’t, or if something goes wrong when it is being executed, the database will report an error.
This result is a data.frame with one row for each entry and one column for each column in the database.
Finally, the last line closes our connection, since the database can only keep a limited number of these open at one time.
Since establishing a connection takes time, though, we should not open a connection, do one operation, then close the connection, only to reopen it a few microseconds later to do another operation.
Instead, it’s normal to create one connection that stays open for the lifetime of the program.
Queries in real applications will often depend on values provided by users.
For example, this function takes a user’s ID as a parameter and returns only the rows with their ID:
library(RSQLite)
connection <- dbConnect(drv = SQLite(),
dbname = here::here("readings","05-database-programming", "data", "survey.db"))
getName <- function(personID) {
query <- paste0("SELECT * FROM Survey WHERE person ='",
personID, "';")
return(dbGetQuery(connection, query))
}
getName("dyer") taken person quant reading
1 619 dyer rad 9.82
2 619 dyer sal 0.13
3 622 dyer rad 7.80
4 622 dyer sal 0.09dbDisconnect(connection)We use string concatenation on the first line of this function to construct a query containing the user ID we have been given.
Database helper functions in R
R’s database interface packages (like RSQLite) all share a common set of helper functions useful for exploring databases and reading/writing entire tables at once.
To view all tables in a database, we can use dbListTables():
connection <- dbConnect(SQLite(),
here::here("readings", "05-database-programming", "data", "survey.db"))
dbListTables(connection)[1] "Person" "Site" "Survey" "Visited"To view all column names of a table, use dbListFields():
dbListFields(connection, "Survey")[1] "taken" "person" "quant" "reading"To read an entire table as a dataframe, use dbReadTable():
dbReadTable(connection, "Person") id personal family
1 dyer William Dyer
2 pb Frank Pabodie
3 lake Anderson Lake
4 roe Valentina Roerich
5 danforth Frank DanforthFinally, to write an entire table to a database, you can use dbWriteTable().
We will always want to use the row.names = FALSE argument or R will write the row names as a separate column.
dbWriteTable(connection, "iris", iris, row.names = FALSE)
head(dbReadTable(connection, "iris")) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosaIn this example we will write R’s built-in iris dataset as a table in survey.db.
Which you can see here:
dbListTables(connection)[1] "Person" "Site" "Survey" "Visited" "iris" We can remove iris as a table with dbRemoveTable() and check it’s been removed with dbListTables().
dbRemoveTable(connection, "iris")
dbListTables(connection)[1] "Person" "Site" "Survey" "Visited"And as always, remember to close the database connection when done!
dbDisconnect(connection)SQL queries with dplyr
In this next section, we will switch datasets for variety sake.
Example: album covers
We will use the
The database represents a “digital media store, including tables for artists, albums, media tracks, invoices and customers”.
From the Readme.md file:
Sample Data
Media related data was created using real data from an iTunes Library. … Customer and employee information was manually created using fictitious names, addresses that can be located on Google maps, and other well formatted data (phone, fax, email, etc.). Sales information is auto generated using random data for a four year period.
The data are saved in our /data folder:
library(here)here() starts at /Users/stephaniehicks/Documents/github/teaching/jhustatprogramming2024list.files(here("readings", "05-database-programming", "data"))[1] "Chinook.sqlite" "survey.db" Connect to the SQLite database
Let’s connect to the Chinook.sqlite file
connection <- dbConnect(SQLite(),
here("readings", "05-database-programming", "data", "Chinook.sqlite"))So we have opened up a connection with the SQLite database. Next, we can see what tables are available in the database using the dbListTables() function:
dbListTables(connection) [1] "Album" "Artist" "Customer" "Employee"
[5] "Genre" "Invoice" "InvoiceLine" "MediaType"
[9] "Playlist" "PlaylistTrack" "Track" I have shown you how to write SQL queries with dbGetQuery().
An alternative approach to interact with SQL databases is to leverage the dplyr framework.
“The
dplyrpackage now has a generalized SQL backend for talking to databases, and the newdbplyrpackage translates R code into database-specific variants. As of this writing, SQL variants are supported for the following databases: Oracle, Microsoft SQL Server, PostgreSQL, Amazon Redshift, Apache Hive, and Apache Impala. More will follow over time.
So if we want to query a SQL databse with dplyr, the benefit of using dbplyr is:
“You can write your code in
dplyrsyntax, anddplyrwill translate your code into SQL. There are several benefits to writing queries indplyrsyntax: you can keep the same consistent language both for R objects and database tables, no knowledge of SQL or the specific SQL variant is required, and you can take advantage of the fact thatdplyruses lazy evaluation.
Let’s take a closer look at the conn database that we just connected to:
library(dbplyr)
src_dbi(connection)src: sqlite 3.46.0 [/Users/stephaniehicks/Documents/github/teaching/jhustatprogramming2024/readings/05-database-programming/data/Chinook.sqlite]
tbls: Album, Artist, Customer, Employee, Genre, Invoice, InvoiceLine,
MediaType, Playlist, PlaylistTrack, TrackYou can think of the multiple tables similar to having multiple worksheets in a spreadsheet.
Let’s try interacting with one.
Using dplyr
First, let’s look at the first ten rows in the Album table using the tbl() function from dplyr:
tbl(connection, "Album") %>%
head(n=10)# Source: SQL [10 x 3]
# Database: sqlite 3.46.0 [/Users/stephaniehicks/Documents/github/teaching/jhustatprogramming2024/readings/05-database-programming/data/Chinook.sqlite]
AlbumId Title ArtistId
<int> <chr> <int>
1 1 For Those About To Rock We Salute You 1
2 2 Balls to the Wall 2
3 3 Restless and Wild 2
4 4 Let There Be Rock 1
5 5 Big Ones 3
6 6 Jagged Little Pill 4
7 7 Facelift 5
8 8 Warner 25 Anos 6
9 9 Plays Metallica By Four Cellos 7
10 10 Audioslave 8The output looks just like a data.frame that we are familiar with. But it’s important to know that it’s not really a data frame. For example, what about if we use the dim() function?
tbl(connection, "Album") %>%
dim()[1] NA 3Interesting! We see that the number of rows returned is NA. This is because these functions are different than operating on datasets in memory (e.g. loading data into memory using read_csv()).
Instead, dplyr communicates differently with a SQLite database.
Let’s consider our example. If we were to use straight SQL, the following SQL query returns the first 10 rows from the Album table:
SQL
SELECT * FROM Album LIMIT 10;Output
1|For Those About To Rock We Salute You|1
2|Balls to the Wall|2
3|Restless and Wild|2
4|Let There Be Rock|1
5|Big Ones|3
6|Jagged Little Pill|4
7|Facelift|5
8|Warner 25 Anos|6
9|Plays Metallica By Four Cellos|7
10|Audioslave|8In the background, dplyr does the following:
- translates your R code into SQL
- submits it to the database
- translates the database’s response into an R data frame
To better understand the dplyr code, we can use the show_query() function:
Album <- tbl(connection, "Album")
show_query(head(Album, n = 10))<SQL>
SELECT `Album`.*
FROM `Album`
LIMIT 10This is nice because instead of having to write the SQL query our self, we can just use the dplyr and R syntax that we are used to.
However, the downside is that dplyr never gets to see the full Album table. It only sends our query to the database, waits for a response and returns the query.
However, in this way we can interact with large datasets!
Many of the usual dplyr functions are available too:
select()filter()summarize()
and many join functions.
Ok let’s try some of the functions out. First, let’s count how many albums each artist has made.
tbl(connection, "Album") %>%
group_by(ArtistId) %>%
summarize(n = count(ArtistId)) %>%
head(n=10)# Source: SQL [10 x 2]
# Database: sqlite 3.46.0 [/Users/stephaniehicks/Documents/github/teaching/jhustatprogramming2024/readings/05-database-programming/data/Chinook.sqlite]
ArtistId n
<int> <int>
1 1 2
2 2 2
3 3 1
4 4 1
5 5 1
6 6 2
7 7 1
8 8 3
9 9 1
10 10 1data viz
Next, let’s plot it.
tbl(connection, "Album") %>%
group_by(ArtistId) %>%
summarize(n = count(ArtistId)) %>%
arrange(desc(n)) %>%
ggplot(aes(x = ArtistId, y = n)) +
geom_bar(stat = "identity")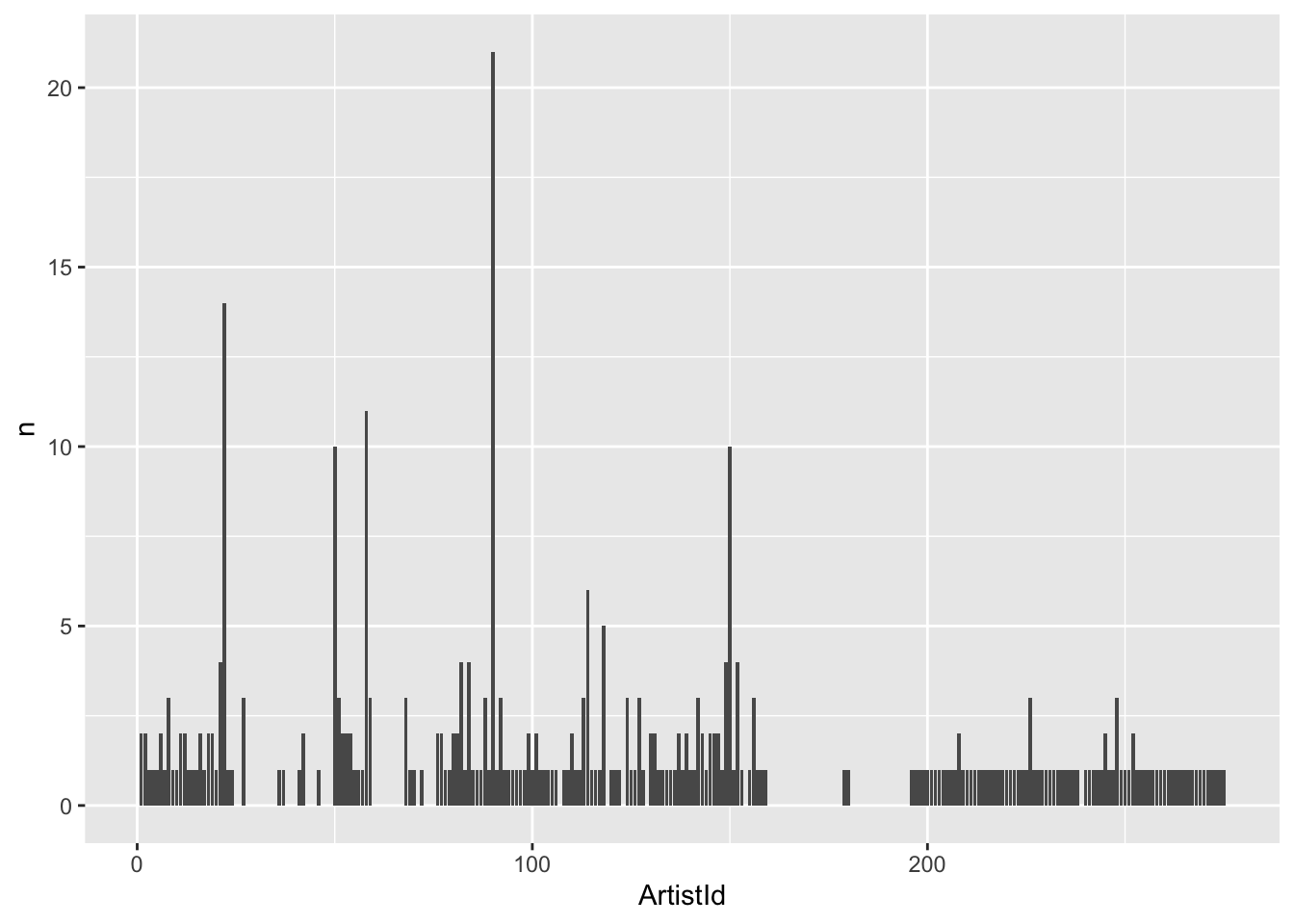
Let’s also extract the first letter from each album and plot the frequency of each letter.
tbl(connection, "Album") %>%
mutate(first_letter = str_sub(Title, end = 1)) %>%
ggplot(aes(first_letter)) +
geom_bar()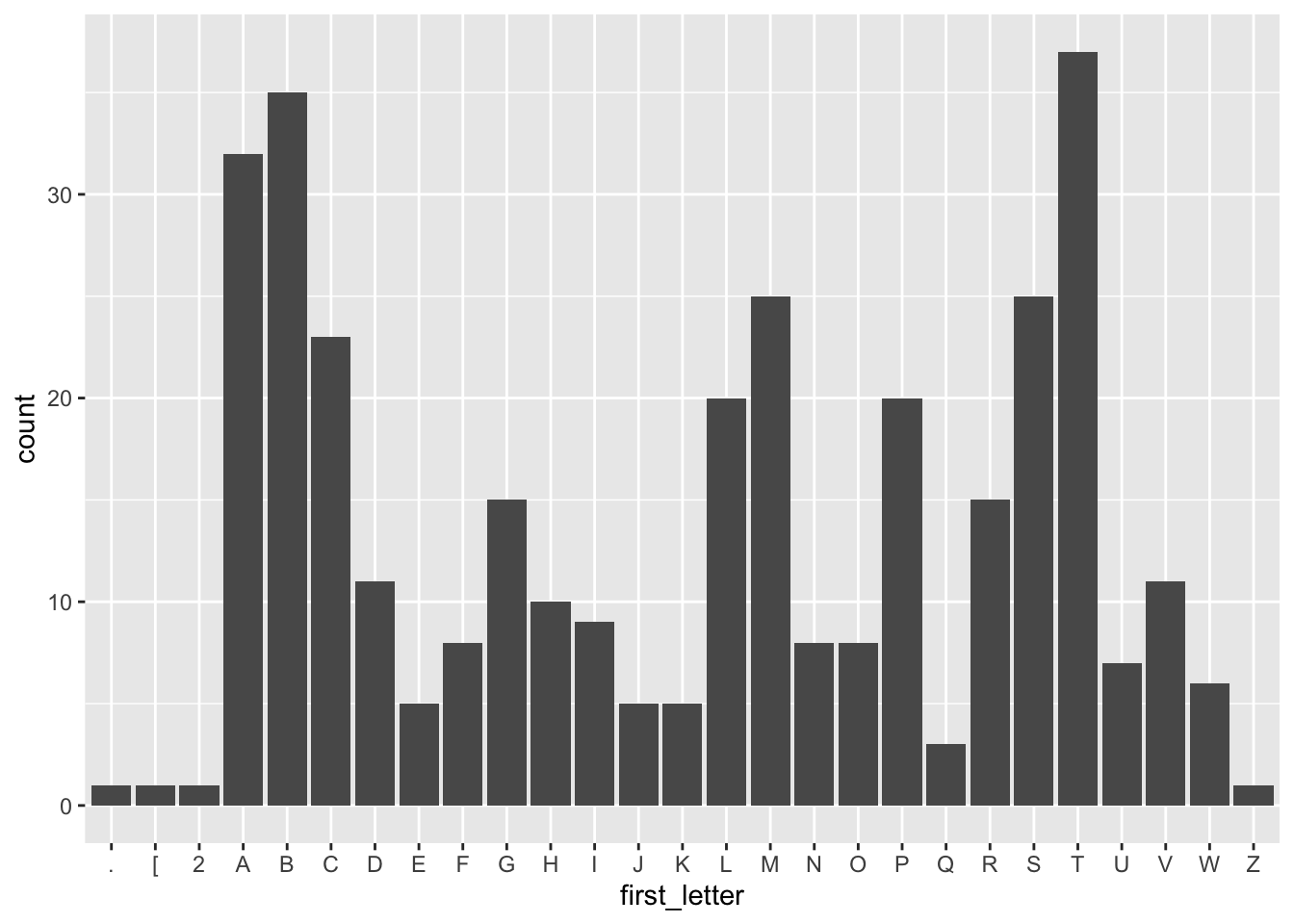
If you decide to make an album, you should try picking a less frequently used letter like E, J, K, Q, U, W, or Z!